4 File Management
4.1 The Essentials
1. Set up a file structure that looks like the following:
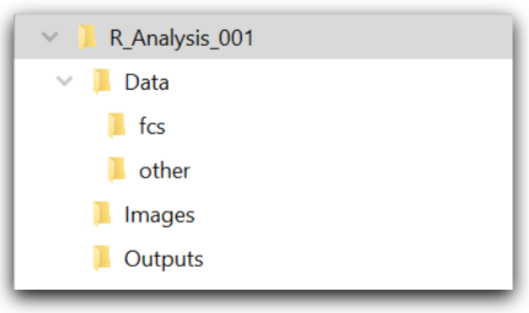
Figure 4.1: The basic structure for analysis keeps everything organized. This structure can be reused for almost any analysis
You can use whichever tool you like. Windows Explorer (or equivalent) is probably be the easiest.
2. Start a R.project in the R analysis folder
- File > New Project > Existing Directory
3. Load the here package:
if(!require(here)) install.packages("here")
#> Loading required package: here
#> here() starts at D:/Analysing-Cytometry-Data-With-R4. Check that the here package “starts” at the right location - it will start where the R Project was set up.
If you need to modify the working directory you can do so with the set_here() function (see below for further details)
5. Download/Copy the tutorial fcs files from here.
6. Copy the downloaded fcs files into the fcs folder and the metadata files into the other folder in your newly created file structure.
4.2 A Deeper Dive
One of the biggest advantages of analysing your data using R is setting up workflows and structures that can be easily reused regardless of the data. This begins with how you set up your file structure and how you move around within that file structure to achieve tasks. During the analysis we will be creating and modifying files and figures so we need to make sure that we are saving them in the correctly and in the correct place.
4.4 The here package
File and fodler structure is an important part of doing an organised analysis. Different packages require different versions of the file path iAs you can see, we need to keep a careful eye on which directory we are working in, while also knowing the full path to each folder and file. This creates a problem. The pathway is specific to the computer you’re working on. This makes it difficult to share code as each person has to change the file paths to their specific needs.
4.5 Set up a folder structure for data and results
We can create folders with:
dir.create(here("Data"))
#> Warning in dir.create(here("Data")):
#> 'D:\Analysing-Cytometry-Data-With-R\Data' already existsWE can create files with
here()
file.create(here("Rtest.txt"))#> Warning in dir.create(here("R_Analysis_001")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001' already
#> exists
#> Warning in dir.create(here("R_Analysis_001/Data")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Data'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Outputs")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Outputs'
#> already exists
#> Warning in
#> dir.create(here("R_Analysis_001/Outputs/flowCut")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Outputs\flowCut'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Images")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Images'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Images/png")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Images\png'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Images/pdf")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Images\pdf'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Data/fcs")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Data\fcs'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Data/Other")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Data\Other'
#> already exists
#> Warning in dir.create(here("R_Analysis_001/Data/RDS")):
#> 'D:\Analysing-Cytometry-Data-With-R\R_Analysis_001\Data\RDS'
#> already exists